Scientists at the IISER Pune assess the stiffness and internal friction of a single folded protein
A new paper published in the Journal of Physical Chemistry Letters from the research group of Dr. Shivprasad Patil investigates the physical properties of proteins in the context of understanding how flexible a protein structure is. Using a special fibre interferometer based Atomic Force Microscope that they developed in the laboratory, the team has been able to make precise measurements of the viscous and elastic components of a single protein domain. This study is anticipated to help understand protein function from a structure standpoint, to monitor small changes in structure (protein conformations), and to obtain a molecular understanding of how some proteins manage multiple functions.
How soft is a single protein ?
Our bodies are swarmed with tiny-little natural nano-machines called proteins performing all the tasks essential for life. To replenish them, we need to eat a protein-rich diet every day. The basic units of these large protein molecules are amino acids, which are stitched together through peptide bonds to form a long chain-like molecule. These large chains fold compactly into a structure. Proteins from our food are processed by our body to cut into single amino acids. These basic units are put together to make different proteins using a delicate machinery inside our cells – like children building new toys out of the same lego pieces again and again.
The compact structures of protein molecules, dictated by its sequence of amino acids, was believed to be entirely responsible for protein function. However, researchers are increasingly realising that this is not the whole story. It is argued that in order to deliver its task, proteins need to be flexible and switch between shapes, which are so little different from each other. For instance, oxygen-carrying hemoglobin has two slightly differing shapes- one when oxygen is bound to it and the other, when it is yet to take up the oxygen. This requires flexibility, which allows proteins to move from one state to another state, the states being referred to as conformations. The ability to switch between shapes effortlessly ( – or with efforts) is likely to determine the protein function and activity. This view is now an active area of research in molecular biophysics.
Protein flexibility, thus needs careful quantification, and falls under the realm of what physicists call study of mechanics. The question is, are proteins rigid yet flexible? – somewhat similar to a clockwork clanking through different shapes each time a chemical signal is received – or perhaps they are soft and flow into a new shape like a delicate dancer. This question is best answered, if we treat protein as a soft complex fluid and measure its rheological response akin to soft materials such as gels, whipped cream, polymer-melts, colloidal suspensions and rubber. The rheology of these materials allows one to quantify liquid-ness and solid-ness of these materials and suggests important time-scales governing their mechanics, such as how long will it take to change its shape fully and the extent to which it is going to change under a given load. Soft materials are subjected to stress-strain cycles to estimate their rheological response.
The challenge in measuring protein flexibility, quantified by its rheological response, lies in its size and extremely small amount of stress developed in it for moderate strains. Atomic Force Microscope, a technique which is able to pick up single protein molecules and apply desired forces on it, is a promising tool for such measurements, yet not without problems of its own. The measurements are performed in liquid environments and the hydrodynamics of the probe moving in liquid itself poses serious challenges and real data gets muddled up with many instrumental artifacts.
“This experimental breakthrough brings many possibilities.” says Shivprasad Patil, the lead researcher of the study.
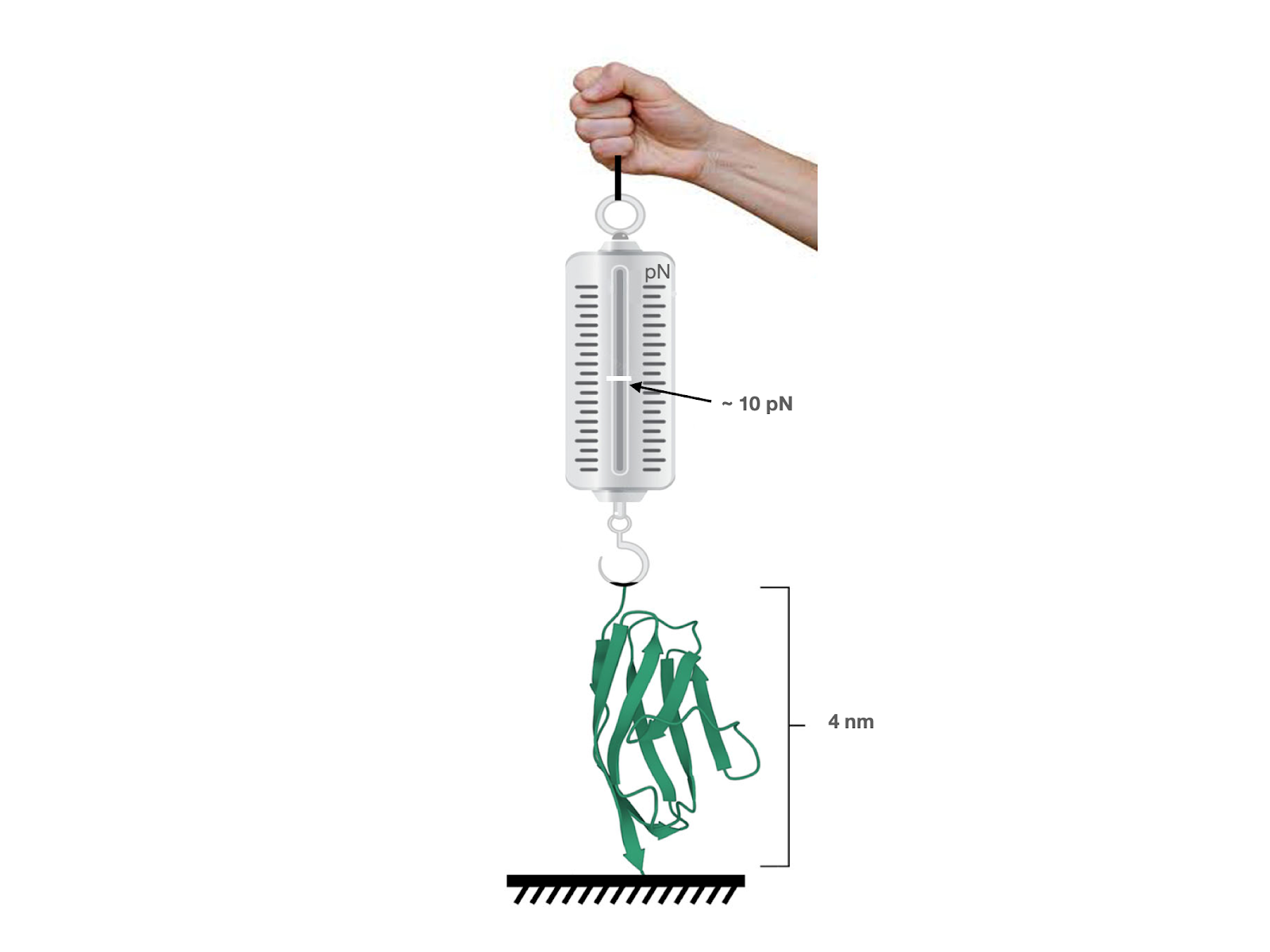
A team of researchers from Nanomechanics lab at IISER Pune, led by Dr. Shivprasad Patil, has now precisely measured the viscous and elastic components of a single protein domain for the very first time. They have developed a special fibre interferometer based Atomic Force Microscope, which, coupled together with their scheme of separating various responses from that of the protein alone, allows them to accomplish such a feat. They tethered their force sensor with single protein domains of giant muscle protein titin.
“We subjected it to an oscillatory load of the order of 10 pN, about 10 billion times smaller than the weight of 1 gm. It produced an oscillatory deformation in the molecule of the order of 10 pm, one billionth of a cm and much smaller than even a hydrogen atom” says Surya Deopa the first author of the study to be published in Journal of Physical Chemistry Letters. It is due to the sensitivity of the interferometer developed by the team that these extremely tiny signals were detected. In short, they have been able to perform rheology on a single molecule by subjecting it to stress-strain analysis, as is done commonly on soft materials.
“This experimental breakthrough brings many possibilities.” says Shivprasad Patil, the lead researcher of the study. If extended to other proteins, this may produce a data repository and allows one to make a direct connection between flexibility and activity. Second, the measurements bolster the view that proteins are dynamic entities residing at a minimum of the free energy with multiple conformational substrates with ever tiny variations in shapes. This also means that protein structures are glass-like. If the origin of protein’s viscous behaviour is probed further, it may allow us a peek into how various shapes (conformations), slightly different from each other, dissipate energy if protein is driven across them in a periodic manner. Such loss in energy could be a useful indicator of how dynamic the protein is, so that a rapid switching between shapes is possible. Such disorders may allow them to perform more than one task, which some proteins are suspected to deliver.”
Reference
Direct and simultaneous measurement of the stiffness and internal friction of a single folded protein. Surya Pratap S. Deopa, Shatruhan Singh Rajput, Aadarsh Kumar, and Shivprasad Patil. Phys. Chem. Lett. 2022, 13, XXX, 9473–9479. https://doi.org/10.1021/acs.jpclett.2c02257
Dr. Shivprasad Patil can be reached on s.patil@iiserpune.ac.in
