Research Summary: Genetic variations in DgoR, a GntR/FadR family transcriptional repressor of D-galactonate metabolism, significantly modulate its repressor function and the metabolic fitness of Escherichia coli strains.

First Author: Dr. Swati Singh is an experienced microbiologist with expertise in bacterial genetics, molecular biology, and biochemistry. She earned her Bachelor’s and Master’s degrees in Microbiology from the University of Delhi. In August 2017, she began her doctoral study in the Department of Biological Sciences at the Indian Institute of Science Education and Research (IISER) Mohali, under the supervision of Prof. Rachna Chaba. Her research focuses on functional and physiological consequences of variations in DgoR, a transcriptional repressor of D-galactonate metabolism in Escherichia coli.
Lab: Prof. Rachna Chaba, Indian Institute of Science Education and Research (IISER) Mohali
Author interview
What was the core problem you aimed to solve with this research?
Sugar acids, the oxidized derivatives of sugars, are widely prevalent in nature and are extensively used as a nutrient source by enteric bacteria. Enteric bacteria degrade sugar acids via the Entner-Doudoroff or Ashwell pathway, which is mainly regulated by GntR/FadR family transcriptional regulators (TRs), characterized by the presence of an N-terminal winged helix-turn-helix (wHTH) DNA-binding domain (Pfam, PF00392) and all α-helical C-terminal effector-binding and oligomerization (E-O) domain, connected by a linker. Although the metabolism of sugar acids, including D-galactonate, is extensively implicated in the colonization and virulence of enteric bacteria, there has been no investigation on the extent of variations in their pathway-specific TRs. Several studies have shown that genetic variations in TRs of metabolic loci can influence host-bacterial interactions by affecting carbon utilization. E. coli DgoR, the TR of D-galactonate metabolism, is the best-characterized GntR/FadR family sugar acid TR in the enteric bacteria. Hence, in the present study, using E. coli DgoR as a representative, we examined E. coli isolates for variations in dgoR and studied their effect on repressor function and impact on the utilization of the carbon source by natural isolates.
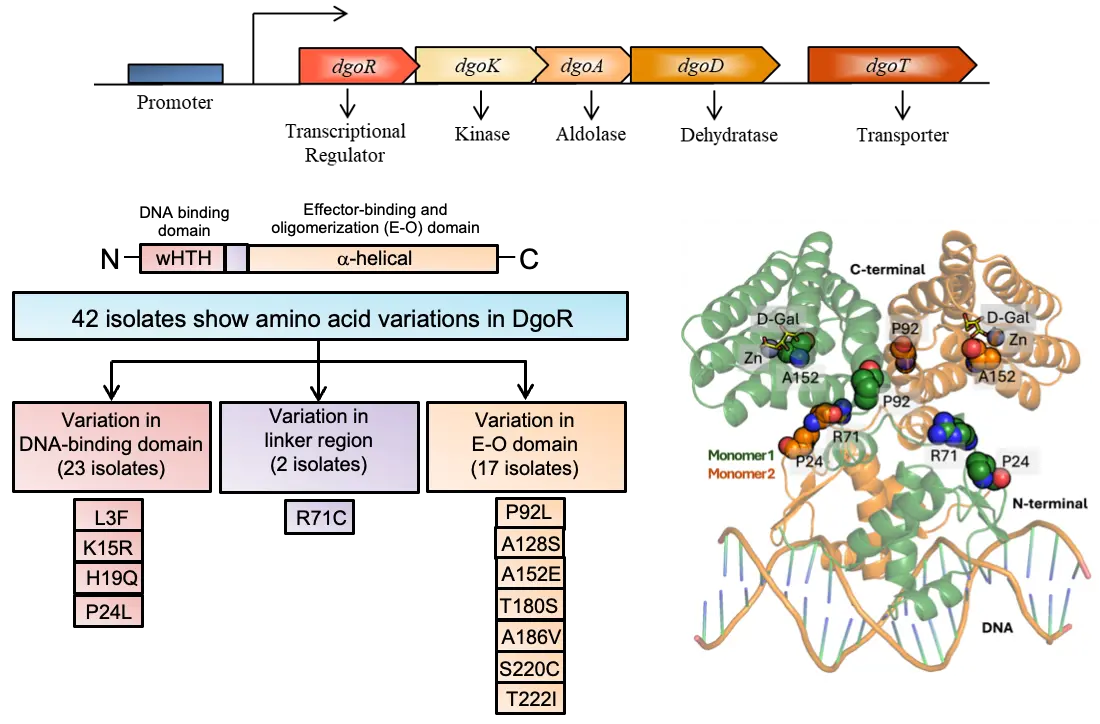
How did you go about solving this problem?
Here, we examined a panel of 340 natural E. coli isolates for variations in dgoR, identified 42 isolates with 12 unique amino acid variations in DgoR, and studied their effect on repressor function. Genetic tests identified variants with a partial loss of DNA-binding ability (P24L and A152E) and variants showing a decreased response to D-galactonate (R71C and P92L). Importantly, corroborating their repressor function, the R71C and A152E variations led to slower and faster growth of natural isolates ECOR-24 and ECOR-39, respectively, in D-galactonate. Biochemical studies showed that, consistent with its compromised inducibility, R71C had a decreased affinity for D-galactonate, and aligning with its reduced repressor ability, A152E had a decreased affinity for the dgo promoter. Because the R71C residue, located in the linker, resulted in a reduced response to D-galactonate, and the A152E residue present in the E-O domain led to a DNA binding defect, we performed simulations to probe their altered allosteric behavior. Molecular dynamics (MD) simulations showed that the correlation patterns, dynamics, and networks of the variants in response to DNA and effector were indeed distinct from the wild type, indicating differences in their structural and functional behavior. Overall, the correlation between in vivo studies, in vitro assays, and MD simulations strengthens the narrative of how specific amino acid variations influence the function of DgoR.
How would you explain your research outcomes (Key findings) to the non-scientific community?
Enteric bacteria such as E. coli can utilize a variety of nutrients as carbon and energy sources, including sugar acids such as D-galactonate. Although utilization of sugar acids has also been implicated in the colonization and virulence of enteric bacteria, the impact of the extent of variations in their pathway-specific TR remains unexplored. Genetic variations in TRs that control metabolic genes can help shape host-microbe interactions by altering carbon source utilization. DgoR, the TR of D-galactonate metabolism, is the best-characterized GntR/FadR family sugar acid TR in enteric bacteria and hence used as a representative in this study. Analysis of a panel of 340 sequenced natural E. coli isolates revealed 42 isolates with 12 amino acid variations in dgoR. We showed that four variants either exhibit decreased sensitivity to D-galactonate (two variations) or lead to a partial loss of DNA-binding ability (two variations). Interestingly, a variation leading to a defect in DNA binding is located in the E-O domain, whereas a variation resulting in reduced sensitivity to D-galactonate is located in the linker region, suggesting compromised allostery as the reason for their altered function. Simulation studies showed that the variations affect the essential dynamics and networks involved in allosteric communication. Notably, genetic studies using gut commensal E. coli strains showed that, indeed, these variations determine the growth of natural isolates in D-galactonate.
In the present study, using a combination of genetics, biochemical, bioinformatics, and simulation studies, we investigated the functional consequences of genetic variations in a GntR/FadR family TR of sugar acid metabolism in E. coli.
What are the potential implications of your findings for the field and society?
In this study, taking E. coli DgoR as a representative, we showed that amino acid variations in sugar acid TRs can affect their function and impact the utilization of these carbon sources by natural isolates. Our study highlights the crucial role of variations in TRs in shaping bacterial metabolism, which in turn defines the host range, preferred ecological niche, and colonization resistance by commensals or disease outcomes of pathogenic infections. Because the ability to utilize scarce nutrients is key for bacterial survival within the host’s complex microbial community, our study stresses the importance of a thorough analysis of variations in sugar acid TR to determine how they influence microbial competition. Such insights could help envision strategies to support the growth of commensals while eliminating their pathogenic counterparts.
What was the exciting moment during your research?
In studies conducted in the 1970s, amongst the various carbon sources tested, the enzymes (DgoK, DgoA, and DgoD) involved in D-galactonate metabolism were shown to be induced in cells grown only in the presence of D-galactonate, suggesting that the D-galactonate pathway is under regulation. Subsequent mutagenesis and genetic mapping studies revealed that mutations affecting the transporter DgoT and the enzymes were located near each other. Additionally, mutations in a putative promoter region—one blocking expression and another causing constitutive expression of Dgo enzymes—were also mapped nearby. These findings suggested that these genes likely form an operon regulated by a TR. Whole-genome sequencing later confirmed that five dgo genes—dgoR, dgoK, dgoA, dgoD, and dgoT—are located adjacent to each other, with dgoR encoding a predicted TR. Although DgoR was long suspected to act as a repressor of D-galactonate metabolism, only in the last few years, detailed studies, mainly published from our lab, have uncovered its regulatory behavior. Initial studies from our lab established DgoR as a GntR/FadR family member and as a transcriptional repressor of D-galactonate metabolism. Our lab further identified the operator and effector of DgoR, investigated its DNA-binding characteristics, and the effector binding cavity. Recently, we proposed the allosteric mechanism that governs the D-galactonate-mediated DNA release from DgoR, which was published in Nucleic Acids Research, followed by our work on the impact of genetic variations in DgoR, which was published in the Journal of Bacteriology, both covering the objectives of my PhD thesis work. Seeing everything come together after so much effort was incredibly rewarding. I am greatly thankful to my PhD supervisor, Prof. Rachna Chaba, for the incredible mentorship and support, my seniors, Dr. Bhupinder Singh and Dr. Garima Arya, whose research work laid the foundation for my project, and all the co-authors who have worked along with me in these projects.
Paper reference: Singh, S., Mishra, R., Kakkar, R.A., Singla, S., Pratap, A., Sharma, G., Sharma, M., and Chaba, R. (2025). Functional consequences of genetic variations in DgoR, a GntR/FadR family transcriptional repressor of D-galactonate metabolism in Escherichia coli. Journal of Bacteriology. https://doi.org/10.1128/jb.00103-25
Explore more
🎤 Career – Real career stories and job profiles of life science professionals. Discover current opportunities for students and researchers.
💼 Jobs – The latest job openings and internship alerts across academia and industry.
📢 Advertise with BioPatrika – Reach the Right Audience, Fast!




