Research Summary: Short- We resolved Myxococcus xanthus DZ2 genome conflicts by removing duplicate-ends, aligning to origin of replication, and circularization, producing a reliable consensus genome. We further classified myxobacterial prophage regions which have adaptive roles in microbial antagonism and ecological interactions.
Longer- The conflicting genome assemblies of Myxococcus xanthus DZ2, a model organism for microbial social behavior, have raised concerns, as two “complete” genomes derived from the same culture stock differ by 6.4 kb while misrepresenting their circular structure. Our computational re-analysis, incorporating duplicate-end removal, alignment to the replication origin, and proper circularization, identified a discrepancy of merely 32 bp, mainly due to sequencing artifacts in homopolymer-rich regions. A reliable consensus circular genome was developed using a draft reference genome assembled by high-quality Illumina data. This study further identified notable diversity in Mx-alpha prophage regions linked to microbial antagonism. Most myxobacteria possess a maximum of two, while DZ2 and DZF1 uniquely retain all three, highlighting their adaptive significance in physiology, development, and ecological interactions.
Author Interview

Utkarsha Mahanta is a DST-INSPIRE SRF at SharmaG_omics Lab at the Department of Biotechnology, Indian Institute of Technology Hyderabad, Telangana, India. Her PhD work majorly focuses on understanding the evolution of myxobacteria in terms of sensing mechanisms, motility strategies, and lethality traits in the phylum Myxococcota, which is carried out under the supervision of Dr. Gaurav Sharma.
Lab: Dr. Gaurav Sharma, Institute: Indian Institute of Technology Hyderabad, Sangareddy, Telangana
Website: https://sites.google.com/view/sharmaglab/
What was the core problem you aimed to solve with this research?
A genome is the blueprint of every life form, and errors in its assembly can lead to significant downstream issues. Being the phylum Myxococcota’s model organism, Myxococcus xanthus is the foundation of several microbiology studies on cooperation, development, and predation. Among its isolates, DZ2 is the most commonly used laboratory strain which was recently assembled by two different labs with almost 6.4 kb difference in their size, questioning their reliability. By resolving these differences using several computational techniques, we provided a reliable consensus genome. Furthermore, though the Mx-alpha prophage region was previously assessed only in M. xanthus, we projected the analysis throughout the phylum, revealing unanticipated diversity and implicating its role in kin discrimination.
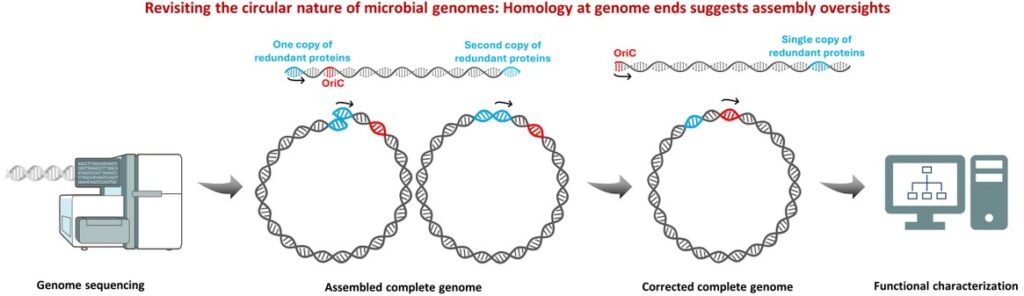
How did you go about solving this problem?
We downloaded all genome assemblies of Myxococcus xanthus DZ2 present in the public space and thoroughly pruned them by deleting duplicated genomic regions present at the genome ends. All assemblies were realigned to begin from the origin of replication (oriC), and conflicts were settled with the help of the previous draft genome, finally yielding a high-confidence consensus genome for DZ2. This improved reference was further contrasted against other M. xanthus strains and other members of the phylum Myxococcota to examine the diversity and distribution of Mx-alpha regions. Our findings demonstrated not only the species-level variability in the number of these prophage regions but also highlighted their role in regulating strain-specific characteristics and ecological strategies of the phylum.
How would you explain your research outcomes (Key findings) to the non-scientific community?
To illustrate my point, consider a poorly made bangle with an overlapping end that could potentially injure the wearer. In the same way, a circular bacterial genome with overlapping sequences can result in flawed analyses and inaccurate predictions, which ultimately hinder scientific progress and societal benefits. By eliminating the defective end of either the bangle or the genome, we can unlock their true potential and make them valuable assets.
A genome recites a beautiful story of its lifeform. By refining DZ2 assembly, we ensured its story can be told with truth and purpose. – Dr. Gaurav Sharma
What are the potential implications of your findings for the field and society?
Our research has important implications both for science and society. With the correction of long-standing inconsistencies in the genome of Myxococcus xanthus DZ2, the best-characterized laboratory strain and key model system for microbial cooperation, development, and predation, we offer a solid reference genome that further grounds the basis for subsequent biological research. An accurate and confirmed genome is important because there are literally thousands of studies that use M. xanthus to investigate core processes. In addition, our identification of unanticipated diversity in Mx-alpha domains in the entire Myxococcota phylum demonstrates how such genetic factors influence microbial behavior, interactions, and fitness. Beyond scientific fundamentals, these findings open applied avenues: myxobacteria are highly productive sources of bioactive natural products, which underpin antibiotic and anticancer drug discovery. Their highly effective degradative enzymes make them promising candidates for biomass conversion, recycling of waste, and bioremediation, while their ecological functions as microbial predators aid in ecosystem reconstruction. Through the integration of genomic accuracy and ecological understanding, this study opens the door to utilizing myxobacteria for drug development, industrial biotechnology, and environmental health.
What was the exciting moment during your research?
The most exhilarating moment in our research occurred when we uncovered a compelling explanation for the substantial variation in genome size between two fully sequenced genomes of the same organism. Through our analysis, we were able to significantly reduce this gap, while still accounting for the minor differences that remained due to sequencing errors. Additionally, we made a significant discovery of critical motility genes situated within the syntenic region of our area of interest. This finding not only highlighted the complexity of genomic architecture but also emphasized the potential implications for understanding organismal behavior.
Paper reference: Creating a consensus genome assembly of Myxococcus xanthus DZ2 by resolving discrepancies between two complete genomes of the same strain and uncovering the Mx-alpha prophage region diversity across phylum Myxococcota. NAR Genomics and Bioinformatics: 2025. https://doi.org/10.1093/nargab/lqaf112
Explore more
🎤 Career – Real career stories and job profiles of life science professionals. Discover current opportunities for students and researchers.
💼 Jobs – The latest job openings and internship alerts across academia and industry.
📢 Advertise with BioPatrika – Reach the Right Audience, Fast!
🛠️ Services – Regulatory support, patent filing assistance, and career consulting services.




