LINE-1 transposon derived R-loops hijack genome stability and drive cancer genome chaos
Research Summary: Active LINE-1 elements form cis and trans R-loops that accumulate in p53-deficient cells, driving genome instability and inflammation, and revealing a novel mechanism of retrotransposon-induced oncogenic stress.
Author interview
Pratyashaa Paul is a doctoral researcher in the laboratory of Dr. Bhavana Tiwari, DBT-Wellcome Trust Intermediate Fellow, at the Department of Biological Sciences, IISER Berhampur. Her research centers on the role of transposable elements in cancer biology. In this study, she spearheaded the investigation into how active LINE1 elements generate RNA-DNA hybrids (R-loops) and promote genomic instability, particularly in p53-deficient cells. Her work led to the discovery of a novel paradigm wherein LINE1 transposons give rise to noncanonical R-loops that destabilize the genome. She played a key role in conducting DRIP-seq experiments, helped develop an RNaseH-GFP based hybrid capture system, and systematically characterized both cis- and trans-acting LINE1 derived R-loops. Pratyashaa’s broader scientific interest lies in understanding how LINE1 derived R-loops drive genome fragility and contribute to human diseases, including cancer.

Lab: Dr. Bhavana Tiwari, DBT-Wellcome Trust Intermediate Fellow, Indian Institute of Science Education and Research Berhampur
Website: https://www.iiserbpr.ac.in/people/profile/btiwari
What was the core problem you aimed to solve with this research?
While most cancer research has focused on mutations in the exonic regions which make up just 1–2% of the genome, the vast majority of the genome, particularly transposable elements (~50%), has remained largely unexplored due to previous technical limitations. With the advent of next-generation sequencing, our understanding of cancer genomics has broadened to include these elements. This study addresses a critical gap: the role of LINE1 (L1) transposons in driving genomic instability and inflammation. Given their ability to mobilize within the genome, L1 elements represent a potent yet under investigated source of DNA damage. By elucidating how L1 intermediates contribute to oncogenesis, this research not only opens new frontiers in understanding cancer biology but also holds promise for identifying novel therapeutic targets.
How did you go about solving this problem?
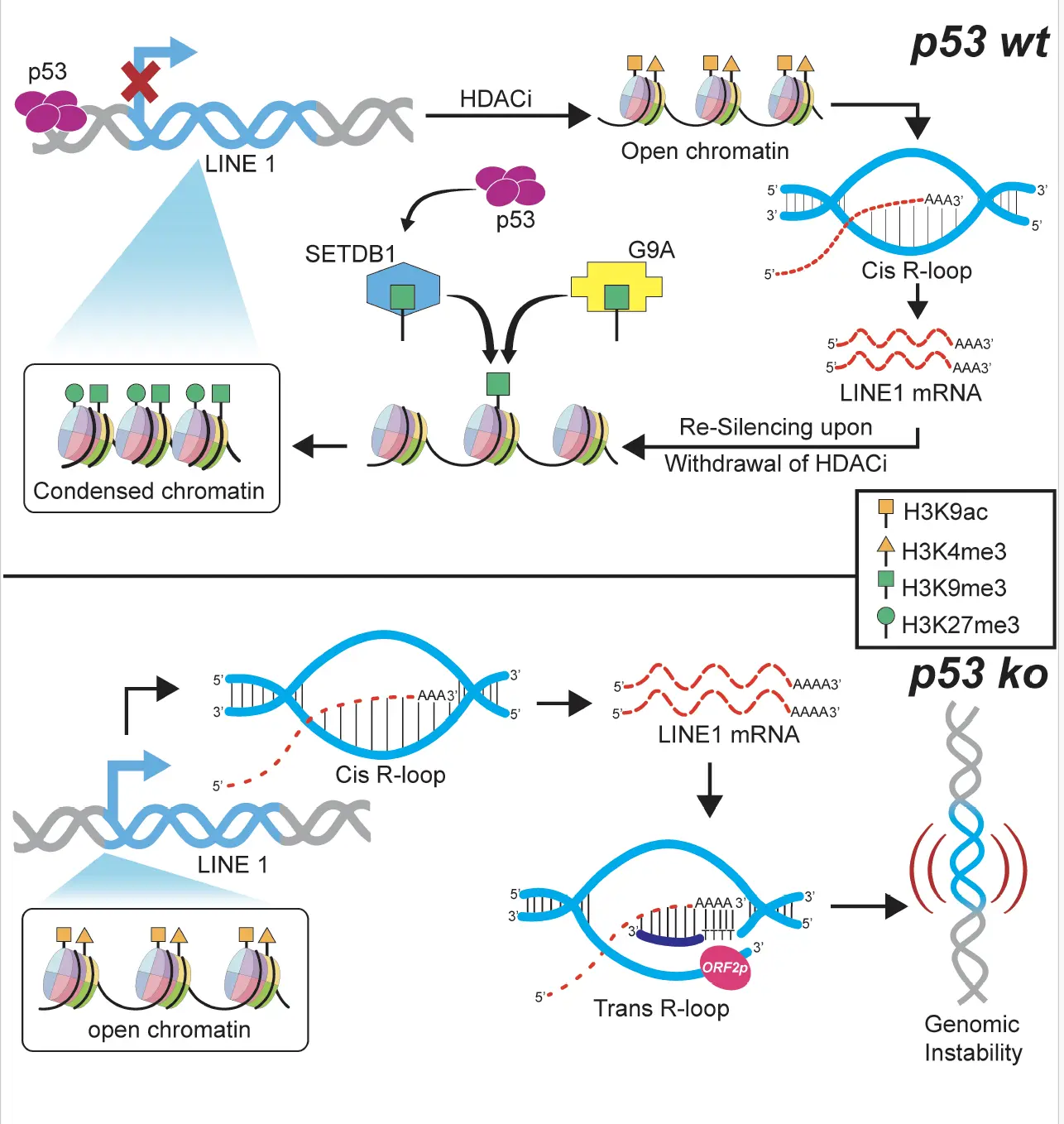
We addressed this problem by integrating DRIP-seq (DNA-RNA Immunoprecipitation Sequencing) leveraging a CRISPR Cas9 engineered p53 knockout model in A375 melanoma cells. This approach enabled a direct comparison of R-loop landscapes between wild-type and p53-deficient cells. DRIP-seq revealed that full-length, retrotranspositionally active LINE1 elements contribute to both cis and trans R-loop formation, with a notable increase in the absence of p53. To validate these observations, we employed immunofluorescence staining and RT-qPCR, which confirmed the accumulation of LINE1 associated R-loops and their association with genome instability and inflammatory signaling. We inhibited LINE1 reverse transcriptase activity, allowing us to isolate the contribution of active retrotransposition to R-loop formation. Additionally, we developed a custom RNaseH-GFP based tool to selectively pull down RNA-DNA hybrids genome-wide, and combined it with LINE1 specific primers to quantify R-loop frequency and distinguish between cis and trans configurations. These data were corroborated by γ-H2AX staining, linking LINE1 derived R-loops to DNA damage in both wild type and p53 knockout cells. This integrated strategy combining sequencing, imaging, molecular tools, and targeted inhibition enabled us to delineate how LINE1 driven cis and trans R-loops promote genome instability and inflammation in the absence of p53.
How would you explain your research outcomes (Key findings) to the non-scientific community?
Nearly half of our DNA is made up of mobile genetic elements, also known as “jumping genes,” a term first coined by Nobel laureate Barbara McClintock. These elements can move around the genome and, if not properly controlled, may disrupt normal genes and lead to diseases like cancer. In our study, we found that when these jumping genes, particularly one called LINE1 become active, they can damage DNA and interfere with the normal regulatory systems of cells. This can create a favorable environment for cancer to grow. By understanding how these mobile elements contribute to cancer development, we hope to pave the way for new strategies in cancer diagnosis and treatment.
What are the potential implications of your findings for the field and society?
Our study uncovers a previously unrecognized mechanism by which LINE1 (L1) retrotransposition intermediates, particularly RNA-DNA hybrids, generate harmful R-loops in p53-deficient cells, thereby contributing to genome instability. These findings redefine how we understand the pathological consequences of L1 activation in cancers with p53 mutations. By establishing the cooperative role of p53, G9A, and SETDB1 in depositing repressive histone methylation marks at the L1 promoter, we not only expand the epigenetic landscape of L1 regulation but also suggest a novel p53-dependent defense mechanism against retrotransposon-driven instability. From a translational perspective, this work has broad implications. First, it provides new biomarkers of genome stress in p53-deficient tumors. Second, our findings offer therapeutic insight: targeting the epigenetic machinery that modulates L1 activity could represent a promising strategy for precision oncology, especially in tumors harboring p53 loss-of-function mutations.
This study uncovers a novel phenomenon where L1 transposition induced R-loops unconventional RNA-DNA hybrids contribute to genome instability in L1 activated cells.
What was the exciting moment during your research?
One of the most exhilarating discoveries was identifying persistent R-loops formed by LINE1 derived RNA-DNA hybrids in p53-deficient cells particularly the trans-acting L1 mRNA-cDNA R-loops, a previously unrecognized phenomenon. Through immunofluorescence imaging, genome-wide DRIP-seq, and targeted qPCR, we established a direct mechanistic link between these retrotransposition-derived hybrids and genome instability. Equally compelling was uncovering how p53 re-establishes silencing of L1 elements after HDAC inhibitor treatment, restoring repressive H3K9me3 marks and preventing activating histone modifications at L1 promoters. This newly described epigenetic feedback loop positions p53 as a critical gatekeeper of transposon activity, revealing fresh insights into its tumor suppressor role.
Paul P, Kumar A, Parida AS, De AK,, Bhadke GV, Khatua S and Tiwari B. p53 mediated regulation of LINE1 retrotransposon derived R-loops. J Biol Chem. 2025;301(3):108200. doi:10.1016/j.jbc.2025.108200 https://www.jbc.org/article/S0021-9258(25)00047-X/fulltext
Explore more
🎤 Career – Real career stories and job profiles of life science professionals. Discover current opportunities for students and researchers.
💼 Jobs – The latest job openings and internship alerts across academia and industry.
📢 Advertise with BioPatrika – Reach the Right Audience, Fast!




