Research Summary: The study uncovered that RNA G-quadruplexes (rG4s) in the 5′ arm of mammalian mirtrons act as structural switches regulating splicing-dependent mirtron biogenesis, revealing a novel post-transcriptional and evolutionary regulatory mechanism.
Author interview

Dr. Uzma Salim is a molecular biologist who recently completed her Ph.D. at IIT Delhi, where she investigated RNA structure–guided regulation of noncoding RNAs, uncovering how RNA G-quadruplexes shape vertebrate mirtron biogenesis.
Linkedin: https://www.linkedin.com/in/uzma-salim-a67b88244/
Twitter: https://x.com/UzmaSalim4
Instagram: https://www.instagram.com/uzma_salim/
Lab: Prof. Vivekanandan Perumal, Indian Institute of Technology, Delhi
Website: https://vperumal.wixsite.com/vivekanandan-perumal
What was the core problem you aimed to solve with this research?
The core problem I aimed to solve was understanding how mammalian mirtron biogenesis—a Drosha-independent, splicing-dependent pathway of microRNA maturation is structurally regulated. While noncoding RNAs are well known for their central roles in gene regulation, most mechanistic studies have focused on canonical miRNAs. As a result, the biogenesis and regulation of non-canonical miRNAs such as mirtrons have remained largely unexplored.
Although mirtrons represent a significant portion of the miRNA population and influence diverse biological processes, the molecular factors governing their precise splicing and maturation were unknown. In invertebrates, mirtron levels are maintained by a TUTase enzyme called Tailor, which specifically recognizes mirtrons via the 3′ splice acceptor site and degrades them to maintain overall mirtron homeostasis. However, no analogous regulatory mechanism has been identified in vertebrates, where mirtrons are more abundant and evolutionarily diverse.
My research addressed this knowledge gap by uncovering a structural mechanism unique to vertebrates. I found that RNA G-quadruplexes (rG4s), specialized secondary structures, are enriched at the 5′ arm of mammalian mirtrons and play a critical role in facilitating their splicing and maturation. Disruption of these rG4s significantly impaired mirtron processing, revealing how RNA structure itself can act as a molecular switch regulating gene expression.
This discovery not only identified the first structural mechanism controlling mirtron biogenesis in vertebrates but also expanded our broader understanding of how RNA secondary structures contribute to post-transcriptional regulation and evolutionary innovation in complex organisms.
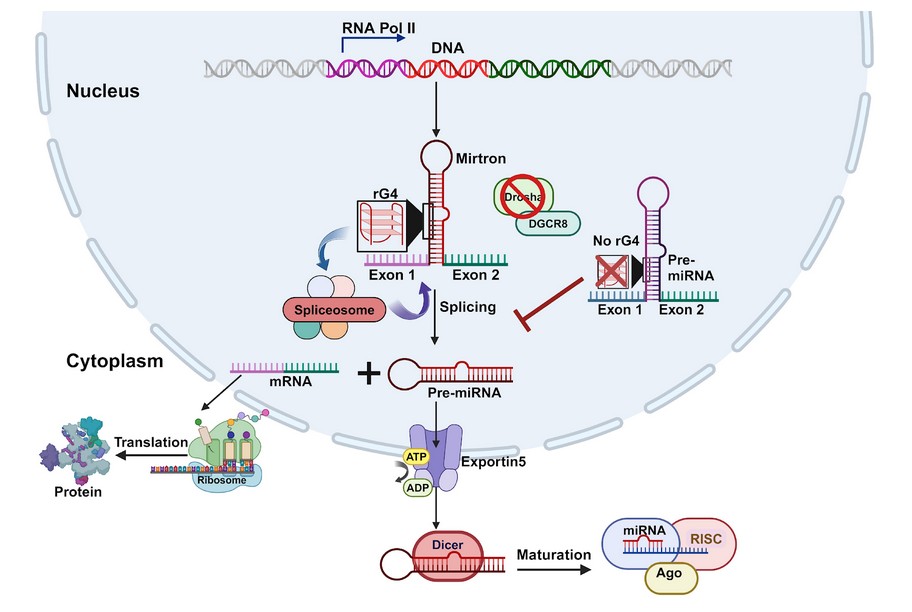
Transcription by RNA polymerase II generates a mirtron-containing transcript that folds into a stem-loop structure featuring an RNA G-quadruplex (rG4) at its 5′ arm. The presence of the rG4 facilitates efficient splicing by the spliceosome, leading to the production of pre-miRNA-like hairpins that bypass the canonical Drosha–DGCR8 processing pathway. These splicing-derived pre-miRNAs are exported to the cytoplasm via Exportin-5, where they undergo Dicer-mediated maturation and associate with Argonaute (Ago) proteins to form the RNA-induced silencing complex (RISC). In contrast, mirtron constructs lacking the 5′ rG4 exhibit inefficient splicing, resulting in reduced pre-miRNA formation and impaired miRNA maturation. In summary, the figure highlights how rG4 motifs function as positive regulators of mirtron processing, underscoring their critical role in non-canonical miRNA biogenesis in mammals.
How did you go about solving this problem?
The study started with a sense of uncertainty. As mentioned, very few studies had focused on mirtrons, and their regulatory mechanisms were largely unexplored. We hypothesized that since mirtron biogenesis depends on splicing, and RNA G-quadruplexes (rG4s) are known to regulate splicing, these structures might also influence mirtron processing. Guided by this idea, we analyzed mirtron sequences for potential structural motifs that could influence their maturation. Interestingly, our analysis identified a strong enrichment of G-rich motifs specifically in the 5′ arm of mammalian mirtrons, which redirected our focus toward understanding how these rG4 structures contribute to mirtron regulation.
The initial phase of the work was quite challenging and involved several technical and analytical roadblocks. We had to test multiple strategies to validate our findings, ensuring that the observed effects were specific to mirtron-associated rG4s and not due to global changes in RNA G-quadruplex motifs. A significant amount of effort went into optimizing cellular assays to accurately assess their functional relevance. Despite these hurdles, through a combination of bioinformatic analysis, biophysical validation, and cellular studies, we demonstrated that 5′ arm rG4s promote efficient splicing and miRNA maturation.
What began as a simple hypothesis gradually evolved into the discovery of a previously unrecognized layer of RNA-based regulation. The journey, though demanding and unpredictable, ultimately proved to be immensely rewarding and provided new insights into non-canonical miRNA biogenesis.
How would you explain your research outcomes (Key findings) to the non-scientific community?
Our genes produce messages in the form of RNA, which act as blueprints for making proteins or regulating other genes. My research focused on a special class of regulatory RNAs called mirtrons, tiny molecules that fine-tune gene activity and are involved in many biological processes, including development and disease.
Unlike conventional microRNAs, which are processed through a well-defined pathway, mirtrons take a detour, they are formed directly from introns, the segments of RNA usually removed during splicing. We discovered that in mammals, mirtron production is controlled by RNA G-quadruplexes (rG4s), unique knot-like structures formed by guanine bases. These rG4s act like molecular switches that enable the cell to splice and mature mirtrons correctly. When these structures are disrupted, mirtron formation is disrupted.
This discovery revealed a completely new way in which cells regulate gene expression, through RNA structures themselves. Interestingly, while simpler organisms use different enzymes for this regulation, mammals appear to have evolved this rG4-based mechanism. Understanding such fundamental processes could, in the long term, guide the development of therapies for diseases linked to RNA processing or gene dysregulation.
What are the potential implications of your findings for the field and society?
The findings from this study have several important implications for both the field of RNA biology and broader biomedical research. Mechanistically, they reveal how RNA G-quadruplexes at the 5′ arm of mirtrons regulate Drosha-independent splicing, adding a new layer of control to non-canonical miRNA biogenesis. This discovery reinforces the idea that RNA secondary structures are not just passive elements but active regulators of gene expression and adds to the growing appreciation of mirtrons as dynamic, evolutionarily flexible components of the transcriptome.
From a broader perspective, understanding mirtron regulation could have far-reaching implications for health and disease. Since mirtrons contribute to the pool of functional microRNAs, their dysregulation could alter gene expression networks, potentially affecting cellular differentiation, development, and disease progression. Identifying rG4s as modulators of mirtron processing also opens new therapeutic possibilities, targeting these structures could enable precise modulation of mirtron-derived miRNAs implicated in diseases such as cancer or neurological disorders.
Overall, this study not only advances our understanding of fundamental RNA structure–function relationships but also lays the foundation for translational research aimed at harnessing RNA elements to fine-tune gene regulation in health and disease.
The study uncovered that 5′ arm RNA G-quadruplexes act as key regulators of mammalian mirtron biogenesis, unveiling a new post-transcriptional regulatory layer. — Prof. Vivekanandan Perumal
What was the exciting moment during your research?
One of the most exciting moments during my research was when, after years of designing and optimizing our constructs, our wild-type and mutant mirtron constructs finally expressed with the expected fluorescence.
Seeing that bright signal was incredibly rewarding—the fluorescence didn’t just illuminate the cells; it lit up my work. It was a powerful moment of validation after years of persistence. From that point onward, the project gained clarity and momentum, and everything began to move in the right direction.
Paper reference: Salim, U., Menon, M. B., Dhamija, S.*, and Vivekanandan, P.* (2025). RNA G-Quadruplexes regulate mammalian mirtron biogenesis. Journal of Biological Chemistry, 301, 3, 108276. https://doi.org/10.1016/j.jbc.2025.108276
Explore more
🎤 Career – Real career stories and job profiles of life science professionals. Discover current opportunities for students and researchers.
💼 Jobs – The latest job openings and internship alerts across academia and industry.
🛠️ Services – Regulatory support, patent filing assistance, and career consulting services.




