Genomic Profiling Sharpens Risk Prediction in B-Cell Acute Lymphoblastic Leukemia
Research Summary: We integrated whole-transcriptome sequencing with conventional genetics to classify B-ALL into clinically meaningful subtypes, improving diagnostic accuracy, identifying novel fusions, and refining prognostic risk stratification in children and adults.
Researcher Spotlight
Jay Singh and Mercilena Benjamin are cancer genomics researchers specializing in RNA sequencing and fusion detection, integrating transcriptomic and genetic data to improve B-ALL classification, risk stratification, diagnostic accuracy, and patient outcome prediction.
Mercilena: Linkedin | Instagram
Lab: Prof. Anita Chopra, Dr. B. R. Ambedkar Institute Rotary Cancer Hospital, AIIMS, New Delhi
What was the core problem you aimed to solve with this research?
A large fraction of B-ALL cases (~25%) remain unclassified using routine cytogenetics, FISH, and RT-PCR, limiting accurate risk assessment and personalized treatment. We aimed to resolve these “B-other” cases and improve prognostic prediction by applying an integrated genomic approach combining whole-transcriptome sequencing with traditional genetic methods.
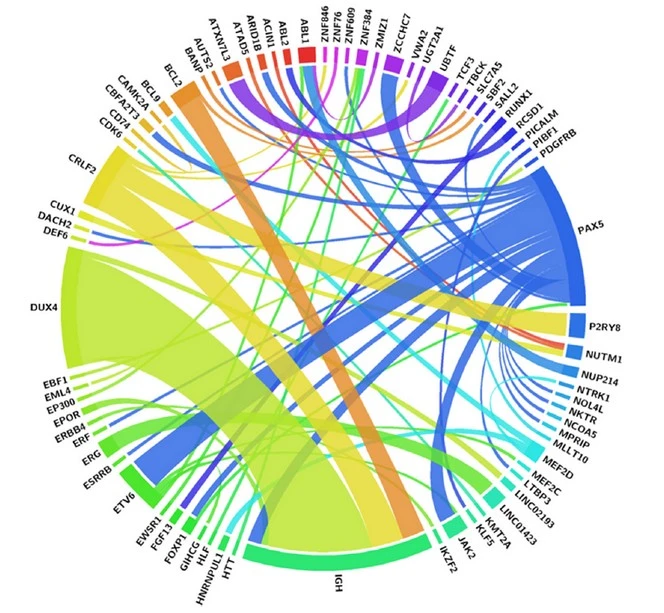
How did you go about solving this problem?
We analyzed 394 B-ALL patients using routine diagnostics (karyotype, FISH, RT-PCR) and performed whole-transcriptome sequencing on 257 samples. Using t-SNE-based gene-expression mapping, fusion detection, mutation analysis, and RNA-seq–derived copy-number profiles, we assigned cases into 23 molecular subtypes and re-stratified patients into clinically meaningful risk groups.
Integrating transcriptomics with routine diagnostics transforms leukemia classification, enabling precise risk prediction and laying the foundation for future precision-guided therapy. — Prof. Anita Chopra
How would you explain your research outcomes (Key findings) to the non-scientific community?
B-ALL which is a blood cancer is not one disease. There are many hidden subtypes that behave differently. Using advanced RNA sequencing, we could read the molecular signature of each patient’s cancer and identify its exact type. This helped classify almost every patient, predict who would respond well to treatment, and discover rare genetic patterns that were previously invisible.
What are the potential implications of your findings for the field and society?
Our integrated approach enables earlier and more accurate leukemia diagnosis, better prediction of treatment response, and personalized risk-based therapy. It can reduce overtreatment in low-risk patients, identify those needing aggressive therapy, and guide the use of emerging targeted drugs. It ultimately improves survival and makes precision medicine feasible in routine clinical settings.
What was the exciting moment during your research?
Discovering several previously unknown gene fusions which includes RUNX1::FGF13 and DUX4::LINC02193 was particularly exciting.These findings reveal entirely new biological mechanisms and may open pathways to targeted treatments for patients who previously had no clear molecular diagnosis.
Paper reference: 10.1016/j.labinv.2025.104201
Explore more
🎤 Career – Real career stories and job profiles of life science professionals. Discover current opportunities for students and researchers.
💼 Jobs – The latest job openings and internship alerts across academia and industry.
🛠️ Services – Regulatory support, patent filing assistance, and career consulting services.





