Research Summary: Cancer treatment often fails due to the cell cycle duration heterogeneities in malignant tumors. We demonstrated that transcriptional memory governs these heterogeneities and predicted ways to improve cancer treatment strategies.
Author interview

Ms. Kajal Charan is pursuing PhD at IIT Bombay in Computational Systems Biology after completing her master’s in Chemistry from MGS University, Bikaner. Her doctoral work focuses on various aspects of oscillatory dynamics in biological systems.
Linkedin – www.linkedin.com/in/kajal-charan98
Twitter – https://x.com/Kajal_Charan27

Lab: Prof. Sandip Kar
University: Indian Institute of Technology Bombay, Mumbai, India
Lab social media: @tsbl_IITBombay on X https://x.com/tsbl_IITBombay
Lab website- https://www.tsbl-skar.com/
What was the core problem you aimed to solve with this research?
The core problem we aimed to address was understanding the origin of tumor heterogeneity, which is a major reason for the failure of cancer therapies. Here, we primarily wanted to unravel how heterogeneities arise in the cell cycle period and phase durations within mammalian cells and especially within cancerous cell lines originating from tumors. To uncover where and how this heterogeneity arises, we focused on the lineage-level correlations of tumor cells – how similar or different lineage pairs are in their cell cycle duration.
In this context, experimental studies have revealed puzzling correlation patterns in cell cycle duration within cell lineage pairs (as shown in the Figure). Surprisingly, daughter pairs remain highly correlated in their cell cycle timings; however, the mother-daughter and cousin-cousin pairs show less correlation in cell cycle duration than the daughter-daughter pair! In this context, one interesting observation is cousin-mother inequality, where cousins have more similar cell cycle duration than mother-daughter pairs. This raises a fundamental question: If the mother isn’t directly correlated with their respective daughters in cell cycle duration, how can cousins end up being more or less similar in cell cycle period to manifest high correlation? To make the matter more complicated, while this inequality is seen in many cell types, sometimes the reverse trend is observed too, where the mother-daughter pair has a higher correlation in the cell cycle duration than the cousin-cousin pair.
These intriguing correlation patterns led us to our broader questions, like:
- How do cells generate such lineage-level correlation patterns in the first place?
- Are these lineage correlations linked to the population-level heterogeneity that drives therapy escape for tumor cells?
- And if we understand the mechanisms leading to such intricate cell cycle duration correlations, could we find ways to reduce tumor heterogeneity and improve therapy outcomes?
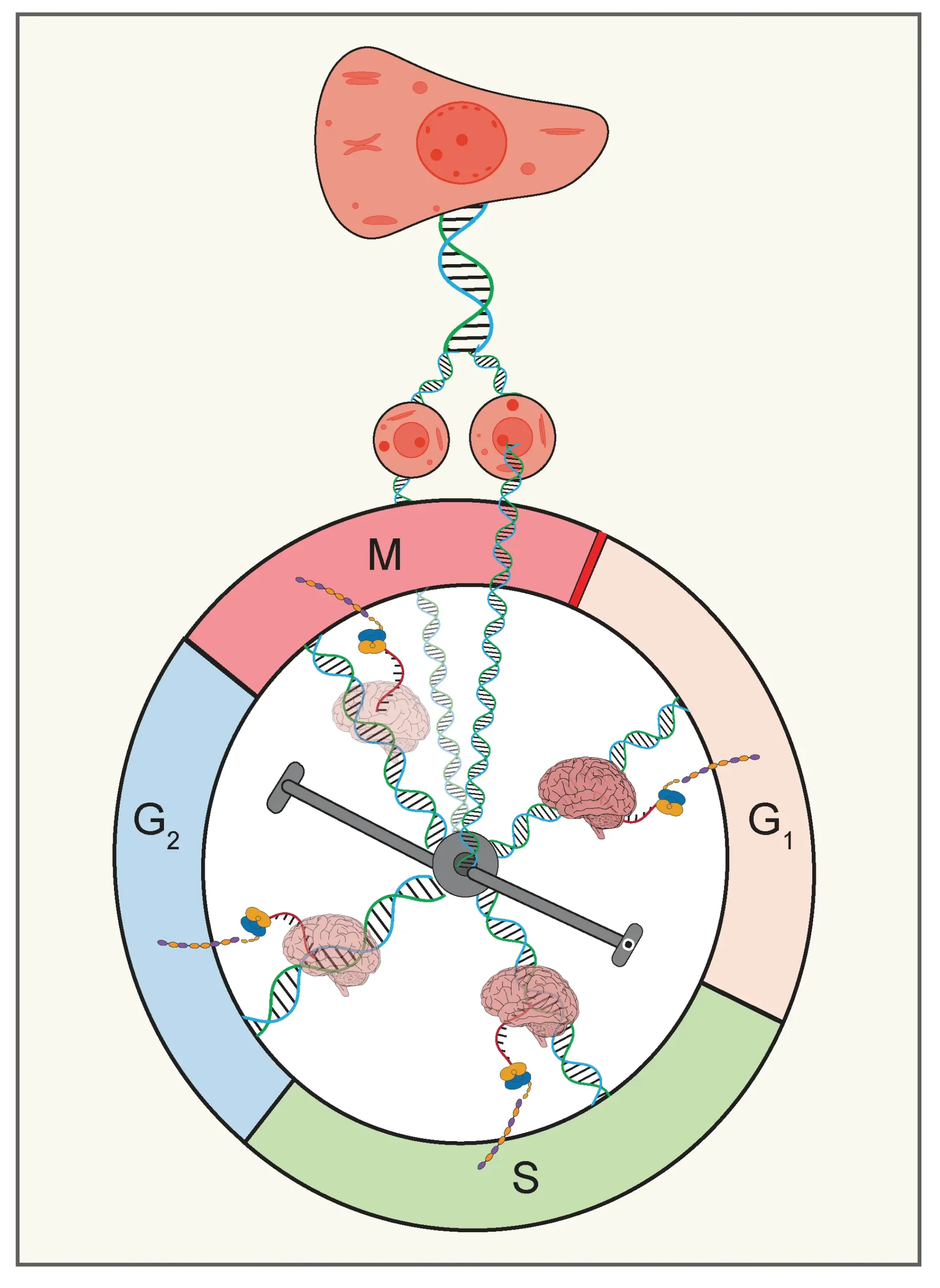
How did you go about solving this problem?
To approach this problem, we first checked whether these lineage correlation patterns—like the cousin-mother inequality – were unique to cancer cells. Interestingly, literature showed that similar patterns exist even in non-cancerous cells and even in bacteria. That suggested there might be a universal regulatory mechanism rather than a specific mechanism driving these correlations.
To start with, we built on a previous experimental study from our group (Govindaraj et al., 2022 ACS Synth. Biol.), which observed cousin-mother inequality in HeLa cells and proposed that transcriptional rate inheritance across lineages could generate such patterns. However, that study couldn’t fully explain the cousin-mother inequality. So, we took this idea further and developed a simplified model to systematically check how varying the extent of transcriptional memory might affect lineage correlations.
To do this, we modeled transcriptional memory using correlated colored noise through the Ornstein-Uhlenbeck process, which allows us to control memory by tuning the noise strength and its autocorrelation time. By systematically varying these control parameters, our simulations revealed an array of possible lineage correlation patterns, including varying extent of cousin-mother inequality observed for various cell types. Moreover, our results showed that these correlation patterns don’t depend on transcriptional memory alone. The correlations can also be influenced by the extent of variability in transcriptional activity, how transcription rates reset during the cell cycle, overall cell cycle duration, and the duration of cell cycle phases.
What causes lineage-level cell cycle duration variabilities remains a long-standing puzzle. Here, we demonstrate that transcriptional memory governs lineage-level heterogeneities. – Prof. Sandip Kar
How would you explain your research outcomes (Key findings) to the non-scientific community?
Cancer therapies often fail because not all cancer cells are the same. A Tumor consists of a mixed population of cells, and some cells in the tumor always find a way to survive. Consider it in this way that you are fighting a group of enemies with a weapon designed to kill them all in the same way. But these enemies are not all alike – some are stronger, some are weaker, and because of this diversity, a few survive even after your attack. Later, those survivors regroup and cause trouble again. This is exactly what happens with cancer. Treatments target the rapidly dividing cells, but slower cells escape the therapy and cause tumor relapse. The big question is: where does this diversity come from if all the cells originally came from the same tumor environment?
To answer this important question, we started looking at the family history of cells in terms of their lifespan over several lineages. This can be quite similar to looking at a family history and analyzing the correlation between siblings, cousins, or parents and children in terms of their lifespan.
We used mathematical modeling to understand how these lineage pairs – mother-daughter, sisters, cousins – show patterns of similarity or difference in their lifespan.
Our model introduced the idea of cellular memory, which gets transferred from mother cells to daughter cells over the generations, and the memory fades away during the cell cycle progression. Think of cellular memory as how much a cell remembers its parents’ behavior to produce mRNA corresponding to any specific protein.
We investigated how the correlation of the lifespan among the lineage pair mentioned above is affected by the time cells spend in particular cell phases (G1 or S-G2-M phases). Imagine our enemy can stay outside the home (like the G1 phase of the cell cycle) and inside the home (like the S-G2-M phase).
Our main finding is that cellular memory (transcriptional memory) plays a significant role in shaping correlations in cell cycle durations, even if there is heterogeneity. If a cell has strong cellular memory, it behaves more like its mother. If it has weak memory, it forgets its mother’s traits, and interestingly, can end up being more like its cousins. This reflects in the cousin-cousin cell cycle duration correlation being higher than the mother-daughter correlation (Cousin-mother inequality phenomenon).
Additionally, we have two interesting findings. First, the sisters consistently remain highly similar in terms of their lifespan because they originate from the same mother, inheriting the same cellular memory from the respective mother cells, and spend a significant amount of cell cycle duration with similar cellular memory.
Second, if cells spend more time at home (i.e., in the S-G2-M phase), they will be more correlated in terms of the cell cycle duration, which means they stay more similar and less diverse. So, in simple terms, if we can make cancer cells stay longer in the S-G2-M phase, they’ll be more similar in terms of their cell cycle progression, and will be easier to target and kill with a specific treatment strategy.
In short, understanding how cellular memory influences cell cycle duration variability allows us to develop strategies that minimize this variability, ultimately enhancing the effectiveness of cancer therapies.
What are the potential implications of your findings for the field and society?
For a long time, it has puzzled scientists why cells across very different cell types – whether bacteria, normal cells, or cancer cells – show these unique lineage correlation patterns. Many explanations have been proposed, but none have fully captured this universal behavior.
Our work provides a generic explanation: these patterns arise from transcriptional memory and how strongly or weakly it is preserved over generations. By dissecting the role of the cell cycle length and its specific phases, we showed that the duration of each phase alters these correlations.
Interestingly, our mathematical model predicts that extending the S-G2-M phase of the cell cycle increases mother-daughter similarity and reduces overall population heterogeneity. Because lower heterogeneity in cancer cells could lead to better therapeutic outcomes. If we can alter how long cancer cells stay in certain phases, we might make them more similar by reducing population heterogeneity – and therefore more receptive to a specific treatment strategy. Thus, our findings provide important insights for improved therapeutic strategies for cancer treatment and have definitive societal relevance.
What was the exciting moment during your research?
The most exciting moment was when our very simple model with transcriptional memory was able to capture diverse features of lineage correlation patterns, including cousin-mother inequality features – something even complex network models had failed to do.
Paper reference: Charan, K. and Kar, S. (2025). Subtle alteration in transcriptional memory governs the lineage-level cell cycle duration heterogeneities of mammalian cells. iScience. https://www.sciencedirect.com/science/article/pii/S2589004225012428?via%3Dihub
Explore more
🎤 Career – Real career stories and job profiles of life science professionals. Discover current opportunities for students and researchers.
💼 Jobs – The latest job openings and internship alerts across academia and industry.
📢 Advertise with BioPatrika – Reach the Right Audience, Fast!
🛠️ Services – Regulatory support, patent filing assistance, and career consulting services.




