Research Summary: We developed a cost-effective targeted nanopore sequencing method to identify HPV integration sites in cervical cancers, revealing functional impacts on nearby cancer-related genes and improving diagnostics potential.
Author interview

Preetiparna Parida is a PhD researcher at MAHE, specializing in viral oncology, nanopore sequencing, and liquid biopsy approaches for HPV-related cervical cancer diagnostics and biomarker discovery.
Linkedin: www.linkedin.com/in/preetiparnaparida
Lab: Dr. Rama Rao Damerla, Manipal Academy of Higher Education
What was the core problem you aimed to solve with this research?
The high cost and complexity of detecting HPV integration events using whole genome or RNA sequencing limit their clinical application. We aimed to create a more accessible method to identify HPV integration sites in cervical cancer.
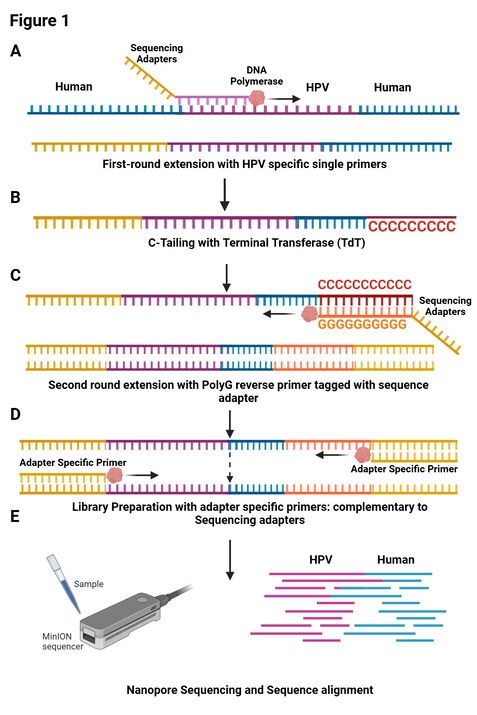
How did you go about solving this problem?
We developed a novel single primer extension strategy to enrich HPV-host chimeric sequences and used nanopore sequencing to detect integration sites. We validated this method using cell lines and tumor samples from HPV-positive cervical cancer patients and compared results with Illumina WGS and Sanger sequencing.
How would you explain your research outcomes (Key findings) to the non-scientific community?
We found a cheaper and quicker way to detect how the HPV virus merges into human DNA in cervical cancer. This helps us understand how the cancer progresses and might help doctors better track and treat the disease.
“This assay marks a pivotal step in translating HPV integration detection into routine clinical diagnostics for cervical cancer.” – Dr. Rama Rao Damerla
What are the potential implications of your findings for the field and society?
Our method offers a low-cost, scalable approach to detect HPV integration in tumors, which can enhance diagnosis, treatment monitoring, and possibly early detection of relapse or disease progression in cervical cancer. It could be expanded to other virus-linked cancers or used to study gene fusions.
What was the exciting moment during your research?
The most exciting moment was when our method accurately detected known HPV integration sites in patient tumors and confirmed their functional impact on neighboring cancer-related genes—proving both the precision and clinical potential of our approach.
Paper reference: Parida P, Mukherjee N, Singh A, et al. Precise identification of viral–host integration events in HPV-positive cervical cancers by targeted long-read sequencing. Tumour Virus Res. 2025;20:200325. https://doi.org/10.1016/j.tvr.2025.200325
More on Cervical cancer and HPV:
- How HPV infection drive cervical cancer https://biopatrika.com/academia/researcher-spotlight/how-hpv-infection-drive-cervical-cancer/
- Part 3: Cervical cancer Prevention https://biopatrika.com/science-society/cervical-cancer-part-1/
- Cervical cancer – Part 2: Staging & Treatment https://biopatrika.com/science-society/cervical-cancer-part-2-staging-treatment/
- Cervical cancer – Part 1 https://biopatrika.com/science-society/cervical-cancer-part-3-prevention/
Explore more
🎤 Career – Real career stories and job profiles of life science professionals. Discover current opportunities for students and researchers.
💼 Jobs – The latest job openings and internship alerts across academia and industry.
📢 Advertise with BioPatrika – Reach the Right Audience, Fast!
🛠️ Services – Regulatory support, patent filing assistance, and career consulting services.




